4 RStudio and Git Setup
4.1 Checking the RStudio environment
4.1.1 R Version
We will use R version 3.6.1, which you can download and install from CRAN. To check your version, run this in your RStudio console:
4.1.2 RStudio Version
We will be using RStudio version 1.2.500 or later, which you can download and install here To check your RStudio version, run the following in your RStudio console:
If the output of this does not say 1.2.500 or higher, you should update your RStudio. Do this by selecting Help -> Check for Updates and follow the prompts.
4.1.3 Package installation
Run the following lines to check that all of the packages we need for the training are installed on your computer.
packages <- c("DT", "devtools", "tidyverse", "ggmap", "ggplot2", "leaflet", "readxl", "tidyr", "scales", "sf", "raster", "rmarkdown", "roxygen2", "broom", "captioner")
for (package in packages) { if (!(package %in% installed.packages())) { install.packages(package) } }If you haven’t installed all of the packages, this will automatically start installing them. If they are installed, it won’t do anything.
Next, create a new R Markdown (File -> New File -> R Markdown). If you have never made an R Markdown document before, a dialog box will pop up asking if you wish to install the required packages. Click yes.
At this point, RStudio and R should be all set up.
4.2 Setting up git
If you haven’t already, go to github.com and create an account. If you haven’t downloaded git already, you can download it here.
Before using git, you need to tell it who you are, also known as setting the global options. The only way to do this is through the command line. Newer versions of RStudio have a nice feature where you can open a terminal window in your RStudio session. Do this by selecting Tools -> Terminal -> New Terminal.
A terminal tab should now be open where your console usually is. To set the global options, type the following into the command prompt, with your actual name, and press enter:
Next, enter the following line, with the email address you used when you created your account on github.com:
Note that these lines need to be run one at a time.
Finally, check to make sure everything looks correct by entering this line, which will return the options that you have set.
Optional:
Check that everything is correct:
Modify everything at the same time:
Set your text editor:
Here nano is used as example; you can choose most of the text editor you might have installed on your computer (atom, sublime, notepad++ …).
Problem with any of those steps? Check out Jenny Bryan Happy git trouble shooting section
4.3 Other Thoughts
4.3.1 Linking git and RStudio
In most of the cases, RStudio should automatically detect git when it is installed on your computer. The best way to check this is to go to the Tools menu -> Global Options and click on git/SVN
If git is properly setup, the window should look like this:
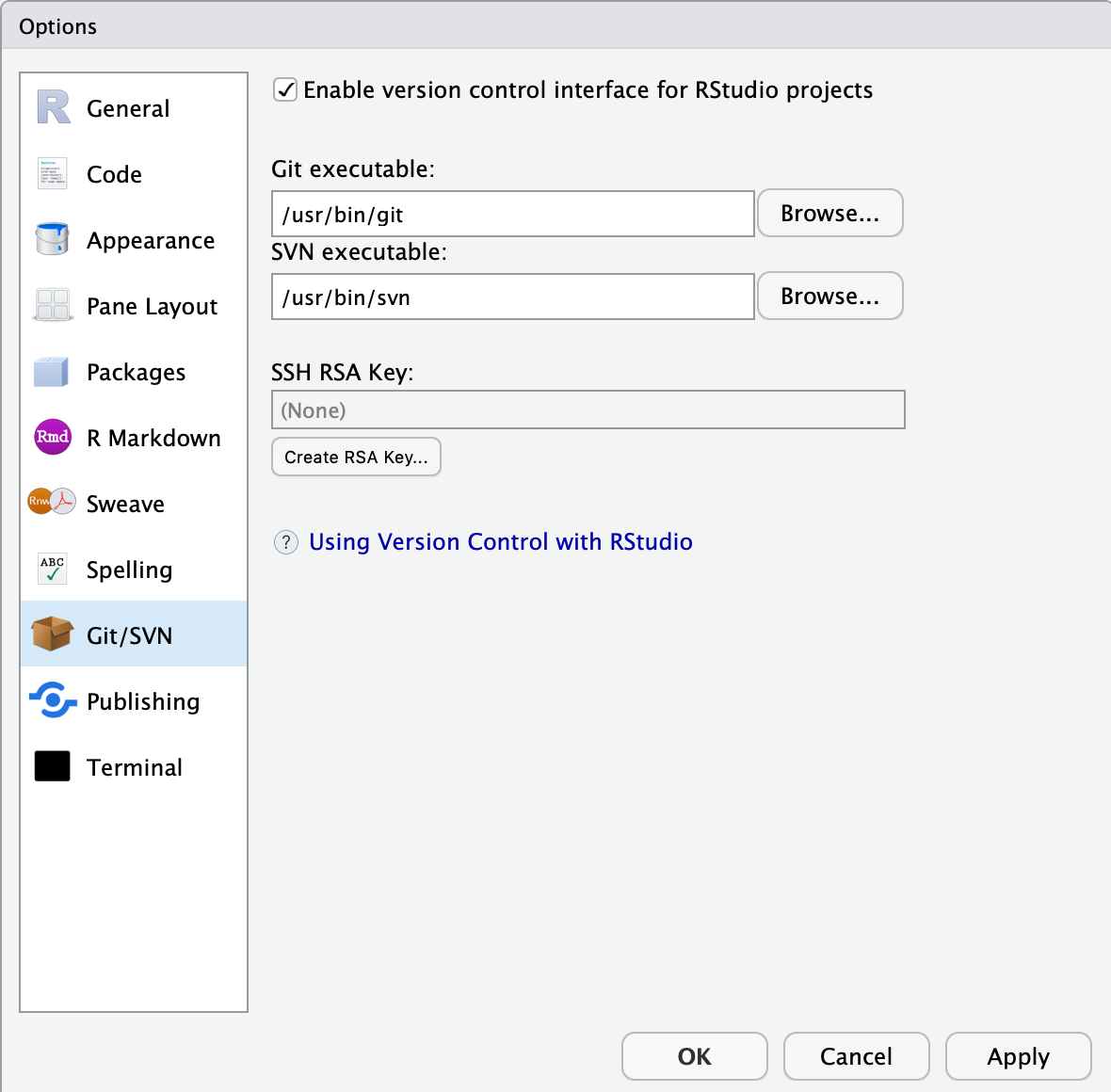
Click OK.
Note: if git was not enabled, you might be asked to restart RStudio to enable it.
4.3.2 Note for Windows Users
If you get “command not found” (or similar) when you try these steps through the RStudio terminal tab, you may need to set the type of terminal that gets launched by RStudio. Under some git install scenarios, the git executable may not be available to the default terminal type.
In addition, some versions of Windows have difficulties with the command line if you are using an account name with spaces in it (such as “Matt Jones”, rather than something like “mbjones”). You may need to use an account name without spaces.
4.3.3 Updating a previous R installation
This is useful for users who already have R with some packages installed and need to upgrade R, but don’t want to lose packages. If you have never installed R or any R packages before, you can skip this section.
If you already have R installed, but need to update, and don’t want to lose your packages, these two R functions can help you. The first will save all of your packages to a file. The second loads the packages from the file and installs packages that are missing.
Save this script to a file (e.g. package_update.R).
#' Save R packages to a file. Useful when updating R version
#'
#' @param path path to rda file to save packages to. eg: installed_old.rda
save_packages <- function(path){
tmp <- installed.packages()
installedpkgs <- as.vector(tmp[is.na(tmp[,"Priority"]), 1])
save(installedpkgs, file = path)
}
#' Update packages from a file. Useful when updating R version
#'
#' @param path path to rda file where packages were saved
update_packages <- function(path){
tmp <- new.env()
installedpkgs <- load(file = path, envir = tmp)
installedpkgs <- tmp[[ls(tmp)[1]]]
tmp <- installed.packages()
installedpkgs.new <- as.vector(tmp[is.na(tmp[,"Priority"]), 1])
missing <- setdiff(installedpkgs, installedpkgs.new)
install.packages(missing)
update.packages()
}Source the file that you saved above (e.g.: source(package_update.R)). Then, run the save_packages function.
Then quit R, go to CRAN, and install the latest version of R.
Source the R script that you saved above again, and then run:
This should install all of your R packages that you had before you upgraded.